Outputs of MIALSRTK BIDS App
Processed, or derivative, data are outputed to <bids_dataset/derivatives>/ and follow the BIDS v1.4.1 standard (see BIDS Derivatives) whenever possible.
BIDS derivatives entities
Entity |
Description |
|---|---|
|
Distinguish different subjects |
|
Distinguish different T2w scan acquisition sessions |
|
Distinguish different T2w scans |
|
Distinguish images reconstructed using scattered data interpolation (SDI) or super-resolution (SR) methods |
|
Distinguish outputs of reconstructions run multiple times with different configuration |
See Original BIDS Entities Appendix for more description.
Note
A new entity id- has been introduced to distinguish between outputs when the pipeline is run with multiple configurations (such a new order of scans) on the same subject.
Main MIALSRTK BIDS App Derivatives
Main outputs produced by MIALSRTK BIDS App are written to <bids_dataset/derivatives>/pymialsrtk-<variant>/sub-<label>/(_ses-<label>/). The execution log of the full workflow is saved as sub-<label>(_ses-<label>)_id-<label>_log.txt`.
Anatomical derivatives
Anatomical derivatives are placed in each subject’s
anat/subfolder, including:The brain masks of the T2w scans:
anat/sub-<label>(_ses-<label>)_run-01_id-<label>_desc-brain_mask.nii.gzanat/sub-<label>(_ses-<label>)_run-02_id-<label>_desc-brain_mask.nii.gzanat/sub-<label>(_ses-<label>)_run-03_id-<label>_desc-brain_mask.nii.gz…
The preprocessed T2w scans used for slice motion estimation and scattered data interpolation (SDI) reconstruction:
anat/sub-<label>(_ses-<label>)_run-01_id-<label>_desc-preprocSDI_T2w.nii.gzanat/sub-<label>(_ses-<label>)_run-02_id-<label>_desc-preprocSDI_T2w.nii.gzanat/sub-<label>(_ses-<label>)_run-03_id-<label>_desc-preprocSDI_T2w.nii.gz…
The preprocessed T2w scans used for super-resolution reconstruction:
anat/sub-<label>(_ses-<label>)_run-01_id-<label>_desc-preprocSR_T2w.nii.gzanat/sub-<label>(_ses-<label>)_run-02_id-<label>_desc-preprocSR_T2w.nii.gzanat/sub-<label>(_ses-<label>)_run-03_id-<label>_desc-preprocSR_T2w.nii.gz…
The high-resolution image reconstructed by SDI:
anat/sub-<label>(_ses-<label>)_rec-SDI_id-<label>_T2w.nii.gzanat/sub-<label>(_ses-<label>)_rec-SDI_id-<label>_T2w.json
The high-resolution image reconstructed by SDI:
anat/sub-<label>(_ses-<label>)_rec-SR_id-<label>_T2w.nii.gzanat/sub-<label>(_ses-<label>)_rec-SR_id-<label>_T2w.json
The slice-by-slice transforms of all T2W scans estimated during slice motion estimation and SDI reconstruction and used in the super-resolution forward model are placed in each subject’s
xfm/subfolder:xfm/sub-<label>(_ses-<label>)_run-1_id-<label>_T2w_from-origin_to-SDI_mode-image_xfm.txtxfm/sub-<label>(_ses-<label>)_run-2_id-<label>_T2w_from-origin_to-SDI_mode-image_xfm.txtxfm/sub-<label>(_ses-<label>)_run-3_id-<label>_T2w_from-origin_to-SDI_mode-image_xfm.txt…
The pipeline execution log file can be found in each subject’s
logs/subfolder as:logs/sub-01_rec-SR_id-1_log.txt
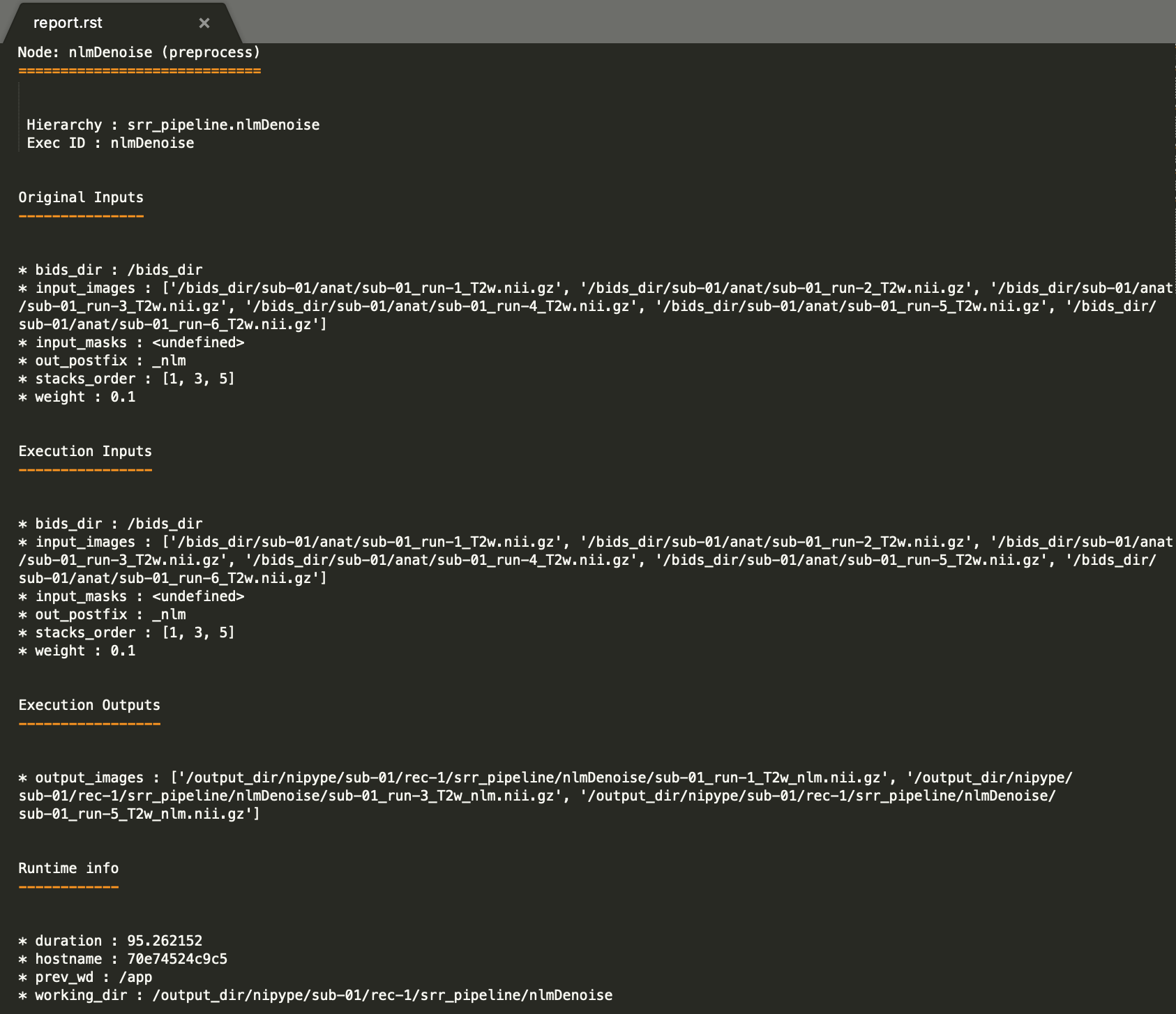
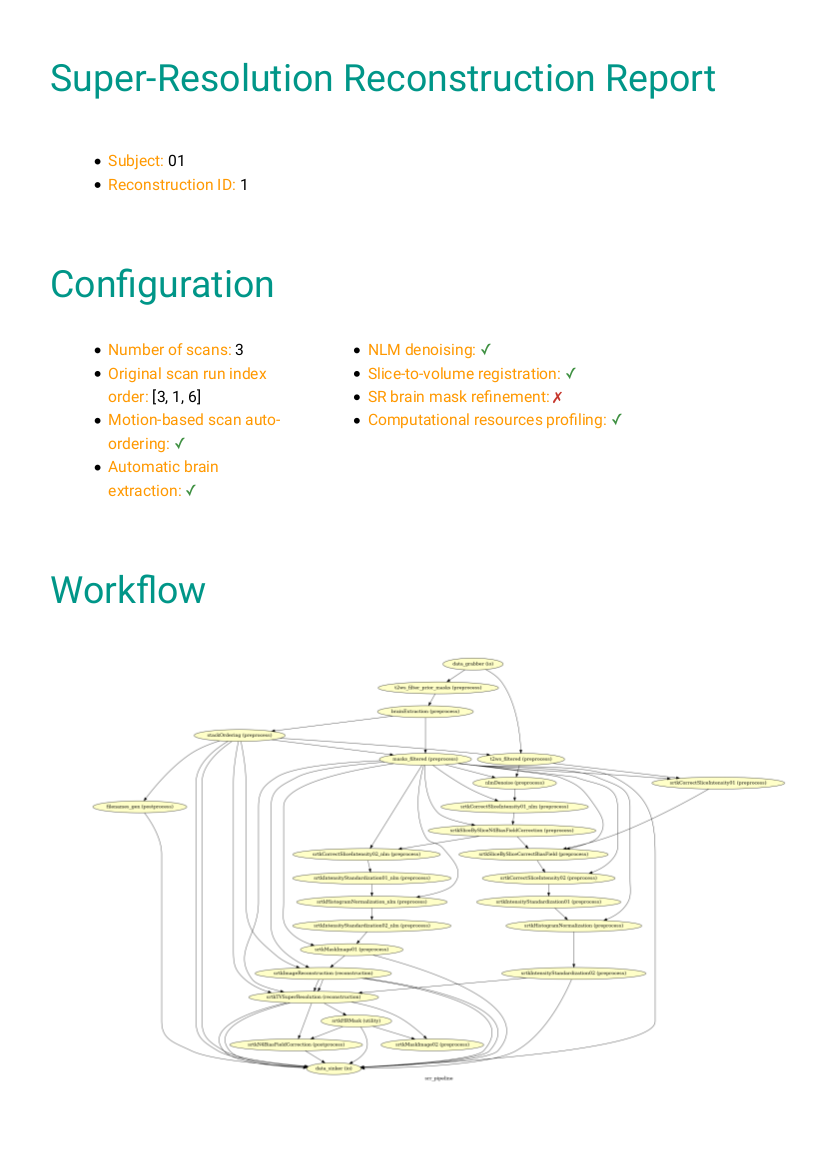
The HTML processing report which provides in one place: pipeline/workflow configuration summary, Nipype workflow execution graph, links to the log and the profiling output report, plots for the quality check of the automatic reordering step based on the motion index, three orthogonal cuts of the reconstructed image, and computing environment summary. It is placed in each subject’s
report/subfolder:The PNG image generated by the stack auto-reordering node and used in the HTML processing report can be found in each subject’s
figures/subfolder as:figures/sub-01_rec-SR_id-1_desc-motion_stats.png
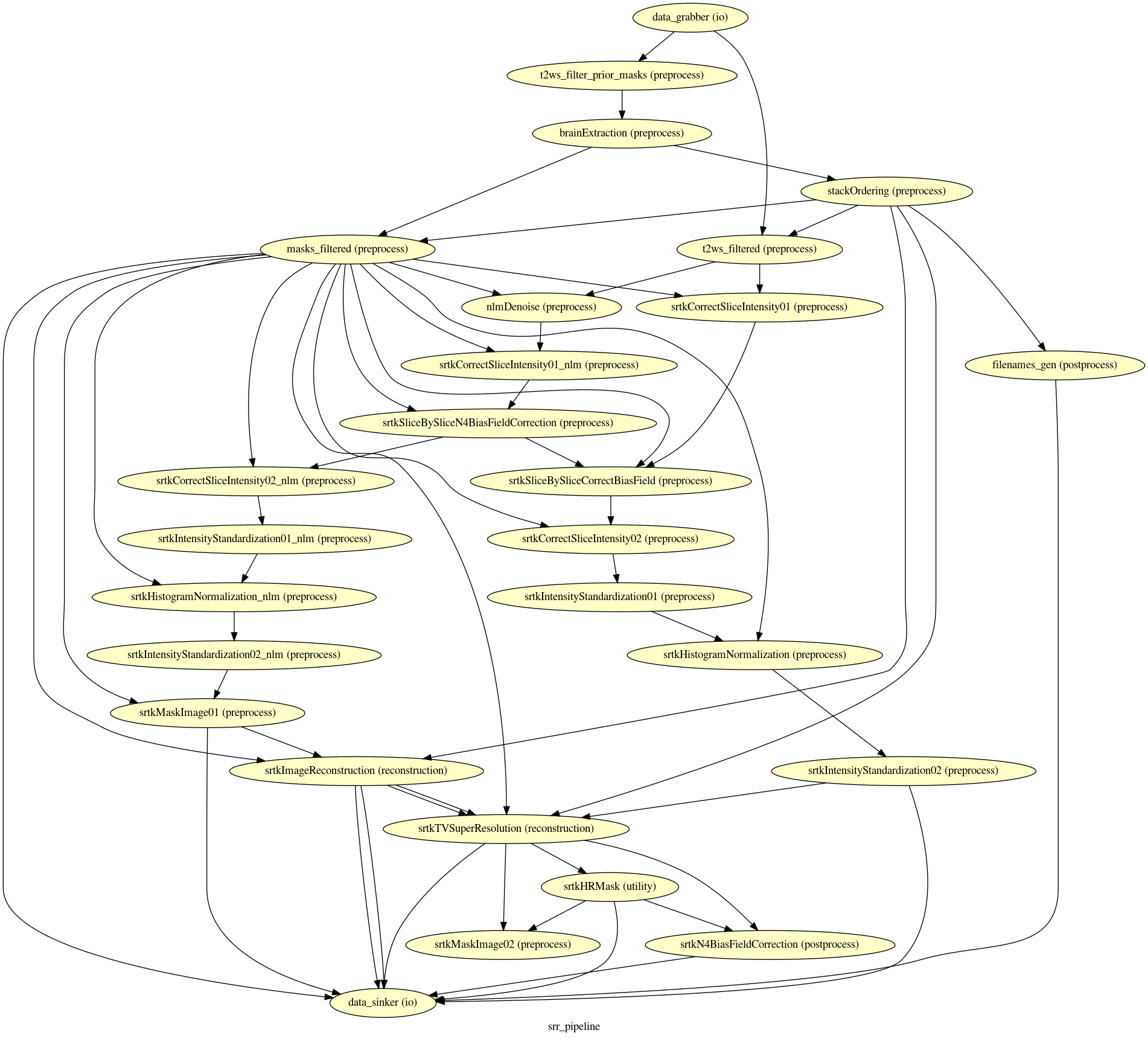
The Nipype workflow execution graph used in the HTML processing report, which summarizes all processing nodes involves in the given processing pipeline, can be found in each subject’s
figures/folder assub-01_rec-SR_id-1_desc-processing_graph.png:
Nipype Workflow Derivatives
The execution of the Nipype workflow (pipeline) involves the creation of a number of intermediate outputs for each subject sub- and each run rec- which are written to <bids_dataset/derivatives>/nipype/sub-<label>/rec-<id_label>/srr_pipeline where <id_label> corresponds to the label used previously for the entity id-:
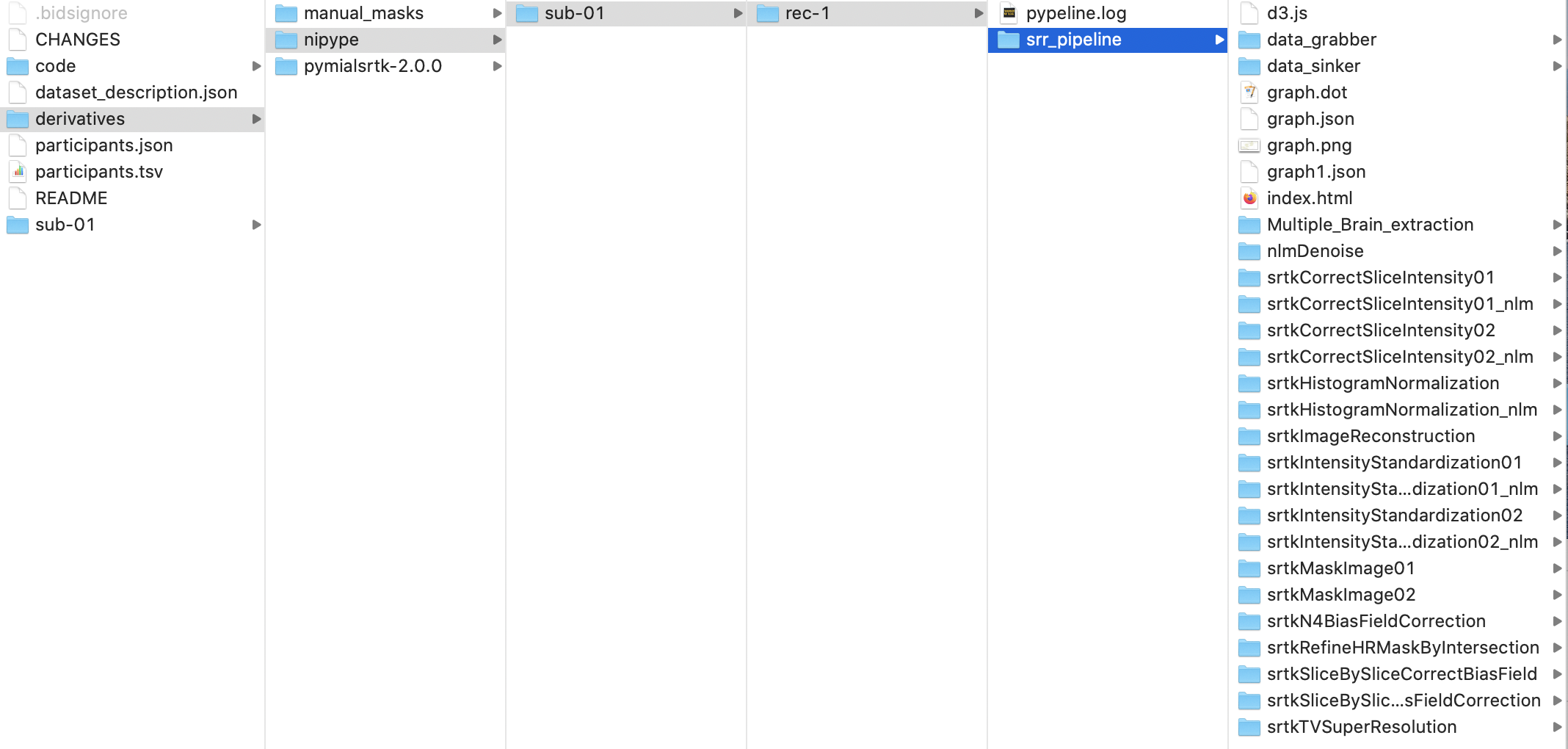
Execution details (data provenance) of each interface (node) of a given pipeline are reported in srr_pipeline/<interface_name>/_report/report.rst